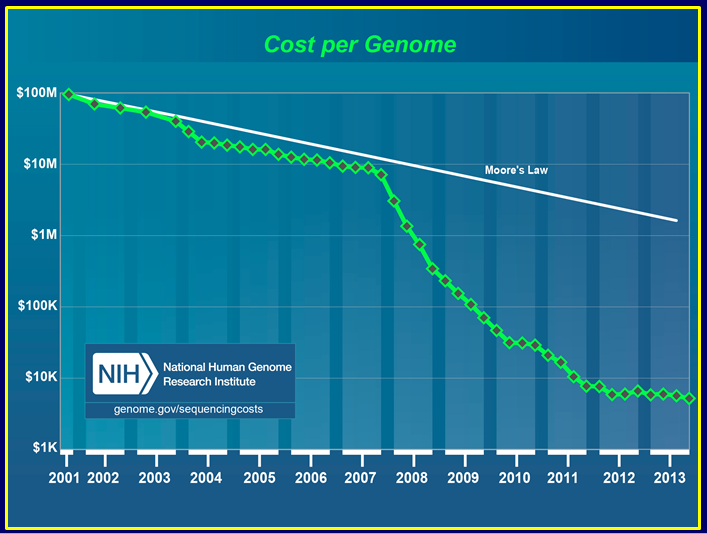
There’s been a lot of attention paid to the tremendous progress in making DNA sequencing so cheap that scanning a person’s genome could cost just $1,000. This pricing free-fall—in contrast to the massive Human Genome Project, which allocated hundreds of millions of dollars to sequencing a single genome in the late ’90s—has occurred markedly faster than with comparable drops for other technologies, such as computers.
Most people would assume that credit is due mostly to the progress made by companies, but in reality the man most responsible for approaching the $1,000 genome is Jeffery Schloss, an unassuming federal employee who works as a program director for the National Human Genome Research Institute.
Schloss launched and oversees the grant programs that have awarded some $220 million to academic scientists and companies developing innovative ways to sequence DNA. Through these programs, NHGRI has invested to some degree in every sequencer on the market—whether directly to the technology developers or indirectly by supporting laboratories that got early access to new techniques and then worked to optimize these new instruments.
The first such funds were issued 10 years ago, and this year the last of the funding will be awarded. Techonomy’s Meredith Salisbury interviewed Schloss recently about how the programs got started, the past decade’s extraordinary achievements, and what is possible today thanks to affordable genome sequencing.
You’re a program director at NHGRI. Aside from helping scientists submit project applications and monitoring the progress of awarded grants, what do you do?
Because we’re talking to lots of different people, we can take a higher-level view of what’s going on in the community. It’s our job to evaluate the opportunities in a research area—what’s getting done and, more importantly, what’s not getting done but could be. If we think there’s a great opportunity, we go back to our agency and recommend it.
Ten years ago, why was driving sequencing costs down a major goal for NHGRI?
At that point people had sequenced the human genome and the genomes of a few model organisms. But there were all these other organisms out there, and having genome sequencing information for them would really advance those fields and also let us learn huge amounts about the human genome.
We had one human genome. The real interest was to see how people who have or don’t have diseases differ from each other in their genomes. We were going to have to sequence very large numbers of genomes, so it couldn’t take multiple years and multiple tens of millions of dollars each time. It was very clear that we needed new sequencing technology, but how to get there was not clear.
So how did the $1,000 genome grant program get started?
We framed it as a 10-year goal. In the planning process someone posed the question, what should be our target for cost? Someone else kind of joked, $1,000. Which of course was ludicrous. Completely ludicrous. It got written into the NHGRI strategic plan, but it was aspirational.
We wanted to reduce costs by four orders of magnitude, which would get you down to about $1,000. We also set an interim goal to drop two orders of magnitude in five years.
Through other grant programs, NHGRI supported most of the academic labs that got early access to new sequencing instruments. Why was that important?
Most of these early adopter labs, particularly large-scale sequencing centers, have a tremendous amount of expertise that helps them implement new technologies. If you look at the cost curve, it was about three years between the time that the first commercial versions of the instruments hit the street and the time that the costs started to drop. Often the platforms come out too early and don’t work very well, so they need to get into the hands of users to find out where they break.
Most companies working on technology development won’t share details with potential competitors, but through this grant program you had everyone speaking at annual meetings. How did that happen?
We thought if we could get people to share as much of their information as possible, they would probably be able to answer questions that others had. Each of them has a lot of knowledge, and they’re really interested in the field advancing. We decided to build that in from the very beginning: if you’re going to get this money, you’re going to have to share information. There has been deep information exchange at these meetings.
How much money has gone toward this drive for cheaper sequencing?
The maximum in any one year was about $20 million, across all the grants. For the last several years it’s been $15 million per year. In total we’ve awarded about $220 million since 2004. That’s the government’s investment. Each of the systems that’s been commercialized has cost in the range of $150 million to $200 million to [bring to market]. We are not funding commercialization of products, we’re funding the research.
Have we reached the $1,000 genome?
We’re approaching it, the goal has not been completely met.
But sequencing is much less expensive now. What has that enabled that wasn’t possible before?
The information you need to make clinical genomics useful comes from doing enough sequencing to figure out the genetic contribution to disease. You have to sequence very large numbers of genomes—thousands of affected people and thousands of controls. That’s happening. You could not do these studies without being able to sequence thousands or tens of thousands of genomes.
On the clinical side, we’ve gotten the cost down to make it possible to do this for individual patients. There are people who have been through a diagnostic odyssey, where no one could figure out what was causing the condition in this family, and people sequenced the family and found genetic changes for which there wasn’t an individual gene test.
The discovery that you could get enough fetal DNA out of maternal blood was really quite astounding. It meant that you could now do a noninvasive genome sequence test. It’s another example of the sort of thing that one can do with this new technology, but it also introduces all sorts of societal and ethical questions.
Talking with the Government’s $1,000 Genome Man
There’s been a lot of attention paid to the tremendous progress in making DNA sequencing so cheap that scanning a person’s genome could cost just $1,000. This pricing free-fall has occurred markedly faster than with comparable drops for other technologies, such as computers. Most people would assume that credit is due mostly to the progress made by companies, but in reality the man most responsible for approaching the $1,000 genome is Jeffery Schloss, an unassuming federal employee who works as a program director for the National Human Genome Research Institute.