A new genomic resource released by scientists promises to streamline genomic testing and precision medicine for patients of all ancestries. It is a significant advance two decades after the first human genome sequence was declared complete.
The original human genome sequence was mainly based on the DNA of a single person, with bits and pieces of DNA from other individuals added over time as scientists learned more about our genomic blueprints. This genome sequence was a reference for the entire genomics community; any DNA sequenced from another person was immediately compared to the connection to glean important insights about disease risk and other traits.
But all along, researchers knew that having a single reference genome would limit what they could learn about other people—especially people whose ancestry isn’t represented by that genome. The genetics-based tests used to guide precision medicine, for example, often can’t find variants that have not been previously identified in the reference genome. Genetic variants common in other populations might be clinically meaningful but will likely go undetected without a more representative reference.

Now, scientists have published what they call a “pangenome” reference. It’s a collection of high-quality sequences from 47 people of diverse ancestries from Africa, Asia, and North and South America. The work comes from the Human Pangenome Reference Consortium, an extensive collaboration of scientists funded to the tune of $40 million by the National Human Genome Research Institute (NHGRI). It’s the first phase of a larger project to sequence 350 people by 2024 to capture a broad range of human genetic diversity.
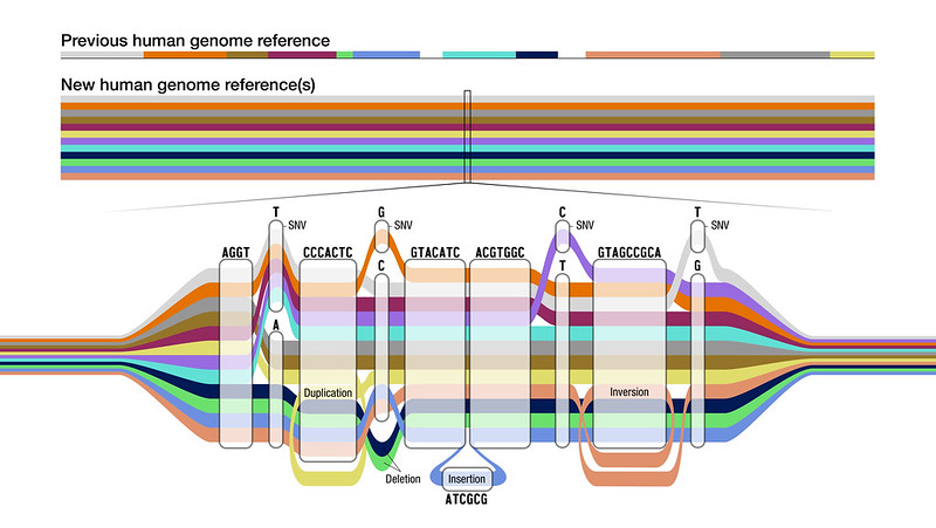
One of the most important outcomes of this new resource should be to improve genome interpretation — the task of spotting clinically relevant genetic variants to diagnose or prevent disease or to match a patient to the proper treatment. “Everyone has a unique genome, so using a single reference genome sequence for every person can lead to inequities in genomic analyses,” said Adam Phillippy, a consortium member and scientist at NHGRI, in a statement announcing the pangenome. Scientists and doctors will have a better chance of finding key genetic variants when comparing someone’s DNA data to this large reference set of genomes than when the reference was just one genome.
“This will help make the reference useful for all people, thereby helping to reduce the chances of propagating health disparities,” said Eric Green, NHGRI director, in the same statement.
The pangenome could also provide a much-needed foundation for new initiatives to automate the genome interpretation process or even to incorporate AI into interpretation. Only now, efforts to streamline the discovery of genetic variants and other critical steps needed to interpret a genome have been constrained by the limited utility of a single reference genome. With nearly four dozen genomes—potentially hundreds by this time next year—it should be much easier to deploy advanced computational tools and improve what is now a largely manual process.










