The Zika virus. Ebola. MERS. If it seems like you’ve been hearing more about infectious disease outbreaks recently, you’re just paying attention. A pair of talks at a recent scientific meeting highlighted evidence that such outbreaks are occurring more often and with more complex infection patterns that make them difficult to treat.
At the annual Advances in Genome Biology and Technology conference, two high-profile scientists—both former fixtures in government science organizations—spoke about trends in infectious diseases. Together, they painted a picture of the rising prevalence of epidemics as well as an increasing challenge: diagnosing and treating people who are infected by multiple pathogens at once. Both expressed hope, however, that advanced studies and improved scientific resources could allow the global health community to respond more quickly and effectively to these threats.
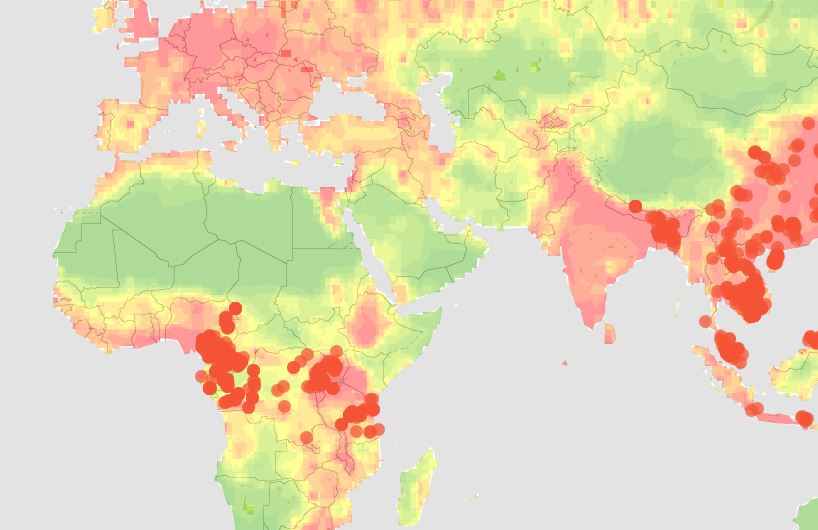
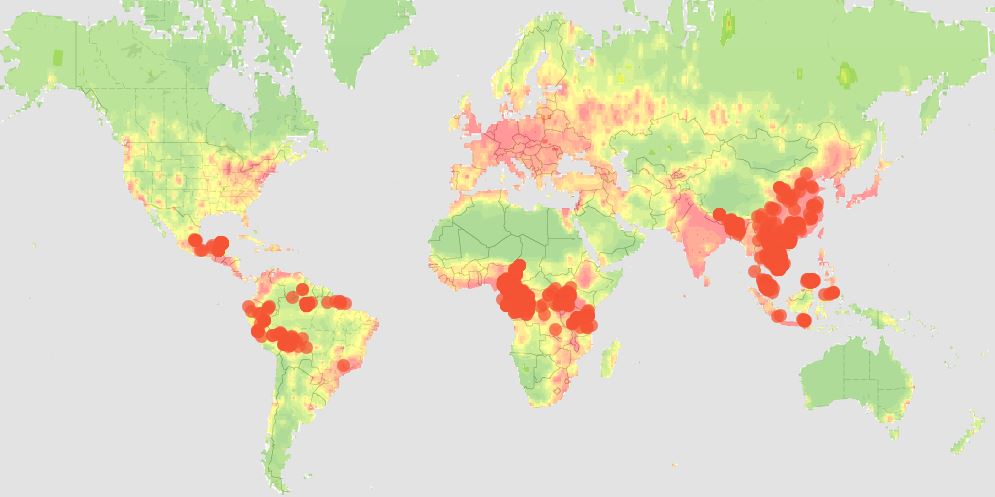
Eddy Rubin, who spent years running the Department of Energy’s Joint Genome Institute and is now CSO of Metabiota, said that the number of viral outbreaks has been increasing during the past two decades. Most of those are zoonotic, or viruses that jump from another species into humans. Rubin said that much of this can be attributed to our rapidly growing population, which keeps spreading into areas previously occupied only by wildlife. With more frequent global travel, outbreaks have a much larger impact than they did before.
Rubin also described efforts to improve our chances of dealing with these viral infections, including the recently launched Global Virome Project. This initiative builds on years-long studies funded by the USAID and aims to identify the viruses most likely to spill over into humans from other species. Scientists are focusing on hotspots—areas where humans live in closest contact to other mammals—and viruses found in multiple species, which indicates that they have already jumped from one organism to another. If successful, the 10-year project will produce a detailed catalog of viruses (most of which are currently unknown) so that future outbreak events can be detected, identified, and responded to far more quickly than is possible today. Ideally, this information could even inform vaccine development programs to immunize against viruses predicted to be the most serious threats to human health.
In a separate talk, former National Science Foundation director and current University of Maryland professor Rita Colwell said that we are now in an “era of polymicrobial infection.” In other words, it’s becoming increasingly common to find patients infected with more than one pathogen at a time. This makes it difficult for diagnostic labs to figure out what’s wrong with a patient using traditional tests, and challenging to treat the patient when each pathogen might require a different medication. Colwell presented data from a microbiome study of more than 70 patients diagnosed with cholera in India, in which scientists found that it was actually a mixture of pathogens rather than just the cholera that sent these patients to the hospital. Results also indicated widespread antimicrobial resistance, suggesting that these co-infections may be even harder to treat. Colwell said that current standards for identifying the cause of a disease need to evolve, and that genomics-based approaches that assess whole communities of microbes instead of tests for a single pathogen will be essential going forward.
While this wasn’t a focus of either talk, doctors have also been reporting more and more cases of patients with infections that are resistant to all known classes of antimicrobial drugs. Clearly, this is a critical time in the battle against infectious disease, and we will need new solutions from genomics and many other fields to get the upper hand.
Infectious Diseases Really Are Out to Get You
Infectious disease outbreaks are occurring more often and with more complex infection patterns, making them harder to treat. While this is a very scary reality, new approaches to diagnosis, treatment and preparation for epidemics hold promise, reported top scientists at a recent conference called Advances in Genome Biology and Technology.